Lettuce is the most valuable, widely consumed, fresh market vegetable crop in the U.S. with an annual farm gate value of ~$3 billion (USDA National Agricultural Statistics Service 2021). L. sativa is an inbreeding diploid (2n = 18) species with limited genetic diversity within cultivated types. Wild species, particularly L. serriola, have been sources of disease resistance genes; however, they remain a rich potential source of variation, particularly for disease resistance, abiotic stress tolerance, and resource use efficiency. The vegetable seed industry depends on public agency plant breeders to provide basic genetic information, new techniques, and relevant, readily usable, advanced lines for their breeding programs.
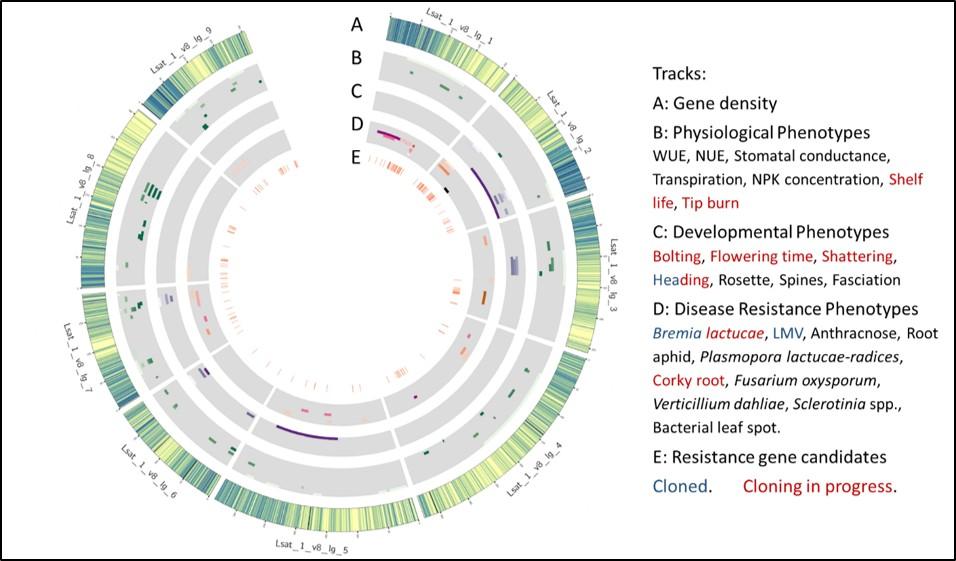
We are developing and applying molecular approaches for lettuce breeding. Lettuce is amenable to classical and molecular genetic analyses. The generation time is usually three to five months, allowing multiple generations each year. We have generated ultra-high density genetic maps for several recombinant line (RIL) populations that have been distributed to the lettuce genetics community. Our on-going genetic experiments have identified over 25 developmental QTLs for a broad range of fourteen horticultural traits. These have been mapped relative to candidate genes involved in germination, leaf development, flowering time, branching, signaling, and sensing in other species; several QTLs were associated with homologs of regulatory genes in Arabidopsis (see figure).
Genomic Distribution of QTLs for developmental, physiological, and disease resistance phenotypes. Circos plot showing the positions of major genes and a subset of QTLs on each chromosome for 24 traits. Each grey bar depicts tracks for groups of experiments.
Our breeding program is focused on the development of advanced breeding lines of all lettuce types with resistance to multiple diseases and improved horticultural performance. Our program emphasizes resistance to multiple diseases including downy mildew, Verticillium and Fusarium wilts, and corky root. We collaborate extensively with other groups to facilitate the development of resistance to other diseases, including a new major emphasis on resistance to Impatiens Necrotic Spot Virus (INSV) and Pythium root rot. We conduct extensive germplasm screens and identified resistant accessions to several of these diseases. We monitor pathogen populations, particularly downy mildew and INSV, in order to select the most effective sources of resistance. We have identified numerous sources of resistance to several diseases in wild species and are introducing resistance from these accessions into all lettuce types using repeated backcrosses. When possible, we aim to introduce genes from a diversity of resistant sources to provide more effective and durable disease control. Our goal is to maximize the evolutionary hurdle for pathogens to become virulent. Tipburn continues to be a priority but in the absence of an efficient screening procedure, resistance can only be evaluated in the later stages of the program in field trials; however, we identified a major quantitative trait locus (QTL), which provides a marker for selection.
We have made many crosses at Davis over the past ~40 years to develop advanced UC breeding lines for all lettuce types with new disease resistance genes. The initial crosses are made at Davis and the early segregating generations are also grown here. This material is then trialed and selected in lettuce-growing areas in collaboration with the USDA and extension personnel in Salinas and Yuma. Resistant germplasm as well as advanced breeding lines are released to the seed industry, so that breeding companies with both large and small research efforts can utilize material generated by our program. This maximizes the diversity of material reaching the growers.
This work is funded by the generous support of the California Leafy Greens Board (CLGRB). Our annual reports to the CLRGB can be viewed at Calgreens – California Leafy Greens Research Board.
