Cultivated lettuce, Lactuca sativa, is a member of the Compositae (Asteraceae) family, one of the largest and most successful families of flowering plants but under-studied relative to several other families. L. sativa is one of the best-studied members of the family and therefore serves as a model for the family. We have been developing genomic resources for Lactuca species within the lettuce genepool and related species in order to facilitate accessing useful genetic variation for lettuce improvement as well as studying genome evolution of this interesting family.
High-throughput DNA sequencing has revolutionized the characterization of natural genetic variation in crop species. We have applied sequencing technologies as they became available to generate ever better genome assemblies (Figure). We have assembled the 2.7 Gb genome into nine chromosomal telomere-to-telomere (T2T) super-scaffolds with few gaps that have been validated genetically; currently, v11 is the latest assembly available in GenBank. We are now completing a ~gapless, T2T hybrid genome assembly based on PacBio and ONT long reads.
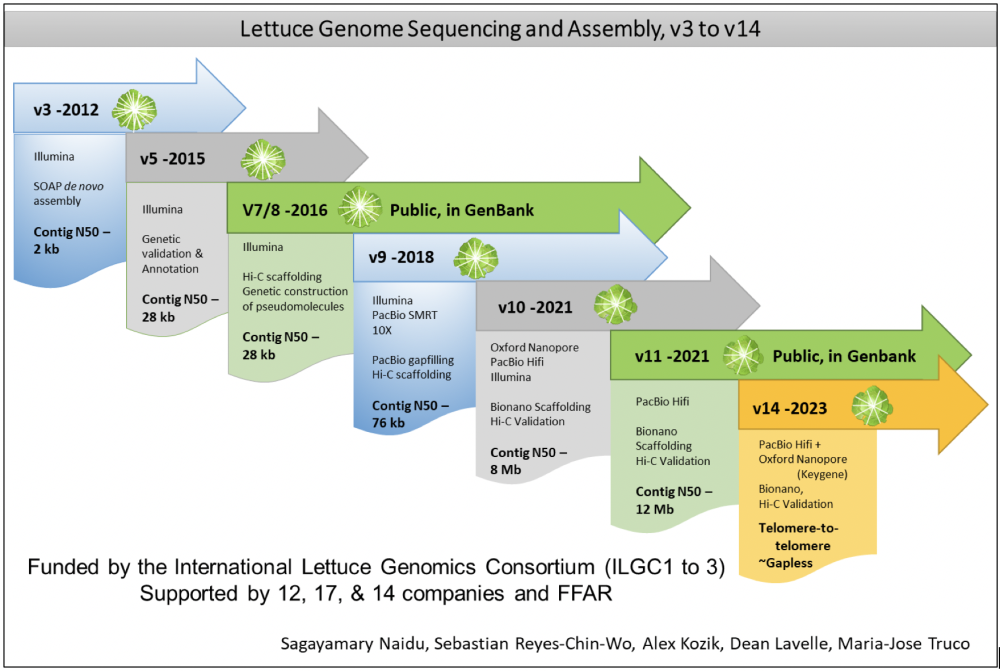
We have also done whole genome shotgun sequencing of ~250 lines in a diversity panel. We are currently focused on generating T2T assemblies of numerous cultivated and wild genotypes, including L. serriola, L. saligna, and L. virosa that comprise the major species in the primary, secondary, tertiary genpools of cultivated lettuce respectively. This is being aided by the acquisition of a Pac Bio Revio DNA sequencer by the UC Davis Genome Center. Extensive genomic tools are now available for lettuce improvement. We are particularly interested in the organization and evolution of clusters of disease resistance genes as well as generating a pan-genome for Lactuca spp.
Paper reporting this work
Reyes-Chin-Wo et al. 2017.
These genome assemblies are the foundation for multiple subsequent papers by us and collaborating labs (see publications).
