
Pathogen Responsible
Rhizorhapis suberifaciens
Description and Sympotoms
Corky root rot was an important disease of lettuce in California and also occurs in Australia, Canada and many countries in Europe. The Gram-negative bacterium that causes corky root, Rhizorhapis suberifaciens (previously known as Rhizomonas suberifaciens; van Bruggen et al., 1988 & 2014), is present in most of the coastal lettuce-growing areas of California. It is a soil-borne pathogen that can affect all lettuce types. Corky root is typically more severe in soils with high nitrate levels and warmer temperatures as well as in fields in which lettuce is grown without rotation with other crops. It was a major problem but is currently of less importance due the use of resistant cultivars and drip irrigation.
Early symptoms of the disease include yellow bands on tap and lateral roots that gradually expand and become green-brown; cracks and pits then develop on the root surface. Subsequently, the entire taproot may turn brown, become severely cracked and non-functional and the lateral root system will be small and damaged. Severely diseased plants wilt in warm temperatures, are stunted, and exhibit generally poor and uneven growth.
Current Management
The use of resistant cultivars is the optimal method for long-term control and lettuce cultivars resistant to corky root are now available. Avoiding over-fertilizing with nitrogen and rotating lettuce with other crops helps reduce CR in susceptible genotypes. CR disease severity may be reduced by using drip irrigation and high, well-drained beds. Additional fertilizer and water may be required to bring an infected crop to maturity.
Genetics of Resistance

Resistance to CR was originally identified in a landrace L. sativa accession PI171669 from Turkey (Dickson, 1963; Sequeira, 1970). This resistance was transferred into several East Coast cultivars including Montello and Green Lake (Sequeira, 1978), South Bay and Tall Guzmaine (Guzman, 1984 & 1986) and subsequently transferred into West Coast cultivars such as Misty Day and Glacier (Ryder and Waycott, 1994).
High levels of resistance to CR are conferred by a single recessive gene designated cor (Brown and Michelmore, 1988). Although additional genes may determine resistance to corky root, genetic analysis and allelism tests have yet to reveal additional loci for CR resistance (Brown and Michelmore, 1988; Mou et al., 2007). Because resistance conferred by the cor gene is recessive and phenotypic evaluation of CR resistance is difficult under greenhouse or growth chamber conditions (Brown and Michelmore, 1988; Michelmore, 2001; M. Govindarajulu, unpublished), CR resistance is an excellent opportunity for breeding using marker-assisted selection.
Genetic Marker Development
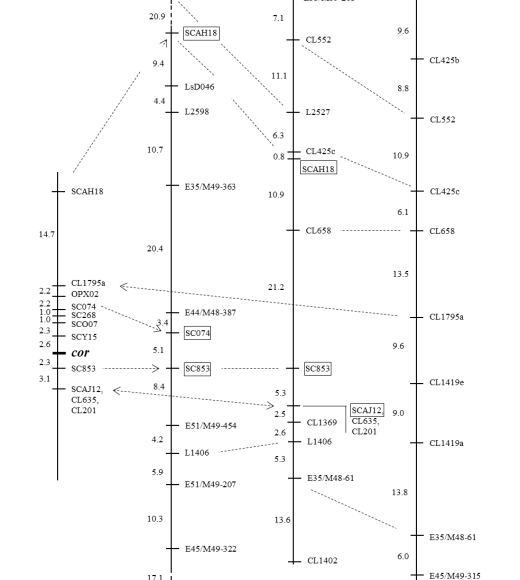
We have identified and developed several co-dominant, PCR-based molecular flanking the cor gene (Moreno-Vazquez et al., 2003). These markers are distributed either side of cor on chromosome 3 (see Figure 1). These can be analyzed without the use of restriction enzymes and are easy and inexpensive to score. At least one of these markers, SCO07, has been used to facilitate introgression of cor into a broad spectrum of lettuce cultivars.
An intra-specific L. sativa map showing the molecular markers located near the cor gene on chromosome 3 (Figure 2). This map from Moreno-Vasquez et al. (2003) was generated using an F2 population from a cross between a crisphead cultivar resistant to corky root ('Green Lake') and a susceptible butterhead cultivar ('Diana'). RFLP markers begin with CL, SCAR markers begin with SC, RAPD markers begin with O. Markers SCO07, SCY15 and SC853 can be used for of cor.
Despite extensive efforts, it proved difficult to identify markers any closer than several cM either side of cor (Moreno-Vazquez et al., 2003). This possibly reflects that cor may be in a chromosomal region of high recombination. Therefore, there is the possibility of recombination between the cor gene and flanking molecular markers. Furthermore, there is a lack of an absolute correlation between resistance and these markers, particularly in romaine types.
We are currently in the process of map-based cloning the cor gene. This has recently provided markers that are tightly linked to cor based on information from the chromosomal pseudomolecules generated as part of the lettuce genome project. High resolution mapping indicated tight linkage to a single scaffold that encoded four genes; these sequences are available gratis for marker assisted selection through a CDA. However, additional analysis indicated that none of these are the causal gene. This page will be updated as new markers for cor are identified and validated.
References
- Bannerot H, Boulidard L, Marron J, Duteil M (1969) Etude de la tolerance au virus de la mosaique de laitue chez la variete Gallega de invierno. Ann Phytopathol 1:219-226
- Blancard D, Lot H, Maisonneuve B (2003) Maladies des salads: identifier, connaitre, maitriser. INRA Editions, Paris, 375p.
- Bos L, Huijberts N, Cuperus C (1994) Further observations on variation of Lettuce Mosaic Virus in relation to lettuce (Lactuca sativa) and a discussion of resistance terminology. Eur J Plant Pathol 100:293-314
- Candresse T, Le Gall O, Mazier M, Maisonneuve B (2006) Virus susceptibility and resistance in lettuce. In: Natural Resistance Mechanisms of Plant to Viruses, eds. Loebenstein G and Carr JP, Springer
- Davies RM, Subbarao KV, Raid RN, Kurtz EA (1997) Compendium of Lettuce Diseases. APS Press, St. Paul, 79p.
- Dinant S, Lot H (1992) Lettuce Mosaic Virus: a review. Plant Pathol 41:528–542
- German-Retana S, Walter J, Le Gall O. (2008) Lettuce mosaic virus: from pathogen diversity to host interactors. Mol Plant Pathol. 2008 9(2):127-36
- Krause-Sakate R, Le Gall O, Fakhfakh H et al. (2002) Molecular and biological characterization of Lettuce mosaic virus (LMV) isolates reveals a distanct and widespread type of resistance-breaking isolate: LMV-Most. Phytopathol 92:563-572
- Le Gall O (2003) Lettuce mosaic virus. “CMI/AAB Description of Plant Viruses” no399. Edited by AT Jones, DJ Robinson, N Boonham and R Mumford. Wellesbourne, UK: Assoc Appl Biol.
- Mazier M, German-Retana S, Flamain F et al. (2004) A simple and efficient method for testing Lettuce mosaic virus resistance in in vitro cultivated lettuce. J Viol Methods 116:123-131
- Nicaise V, German-Retana S, Sanjuan R, Dubrana MP, Mazier M, Maisonneuve B, Candresse T, Caranta C, LeGall O (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol 132:1272-1282
- Ryder EJ (1979) Vanguard 75 lettuce. Hortsci 14: 284-286
- Ryder EJ (1991) Salinas 88 lettuce. Hortsci 26: 439-440
- Ryder EJ (1970) Inheritance of resistance to common lettuce mosaic. J Am Soc Hort Sci 95:378-379.
