The lab has several research objectives:
- Comparative and functional genomics of disease resistance genes in plants.
- Classical and molecular genetics of lettuce and its pathogens.
- Development and deployment of novel technologies for crop improvement.
- Genomics of oomycete pathogens that cause downy mildews.
- Genomics of understudied crop species.
Plants express hundreds of disease resistance genes. A major focus of our research is the comparative and functional genomics of disease resistance, particularly in lettuce. Our investigations involve the large-scale genomic analysis of resistance gene evolution and function as well as comparative genomics analysis of resistance across multiple species. These studies revealed that some resistance genes are not evolving as fast had been previously assumed and led us to a model for the evolution of resistance genes. An ultimate aim of our studies is to be able to direct the evolution of resistance genes ex planta to recognize new pathogen ligands as well as to design resistance genes to encode new recognition specificities.
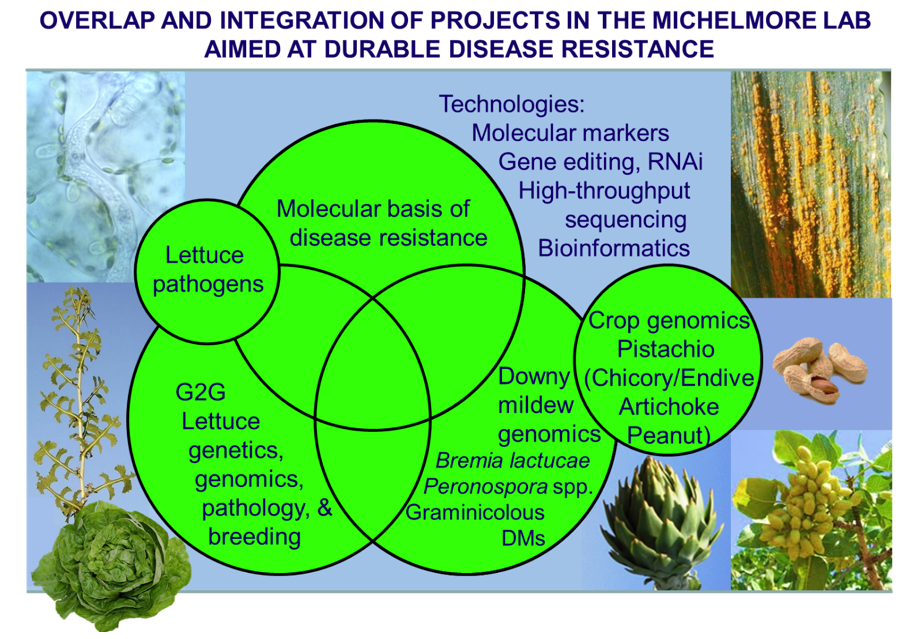
Studies on lettuce include classical genetics of disease resistance, tolerance to abiotic stressors, water and nitrogen use efficiencies, genetic dissection of these traits using molecular markers, large scale genome sequencing, comparative genomics, transgene expression and genome editing, and characterization of resistance gene clusters at the molecular level. We utilize a combination of top-down (genetic) and bottom-up (candidate gene) strategies to clone genes for disease resistance and other horticulturally important traits. A long-term goal of the lab is to deploy stacks of effective resistance genes, so as to maximize the hurdle for pathogens to evade recognition and so increase the durability of resistance.
Our classical genetic studies of lettuce involve screening lettuce germplasm for resistance, determining its genetic basis, introgression of resistance into cultivated genotypes, and release of advanced resistant lines. Most of our efforts have focused on resistance to downy mildew (Bremia lactucae), corky root (Rhizorhapis suberifaciens), and wilts caused by Verticillium dahliae and Fusarium oxysporum. We recently expanded our efforts to include resistance to Impatiens Necrotic Spot Virus (INSV) and Pythium uncinulatum because of their increasing impact on lettuce production. These classical studies provide genotypes for our molecular investigations as well as advanced breeding lines for the commercial breeding industry. In parallel, we also continually monitor variation in B. lactucae and recently INSV to assess the variation in virulence and direct the deployment of effective resistance genes.
Over the years we have deployed a broad array of molecular marker and sequencing technologies as they became available, contributing to their development in several cases. The lab is located within the UC Davis Genome Center, which provides us with access to the latest technologies. We developed detailed genetic maps of lettuce and demonstrated that genes for resistance to diverse pathogens are clustered in the genome indicating a common origin and function. We sequenced and assembled the genome of L. sativa cv. Salinas, first using Illumina sort reads in collaboration with the BGI (Shenzen, China) and subsequently using Oxford Nanopore long reads and PacBio HiFi reads. We now have a telomere-to-telomere assembly of the lettuce genome. We are currently generating additional high-quality assemblies of cultivated and wild genotypes using short and long reads with the goal of generating a pan genome for lettuce.
We have developed stable and transient transformation as well as genome editing protocols for lettuce. However, while it is relatively easy to introduce DNA into lettuce, transgenes are often not consistently expressed in stable transgenics due to silencing. Therefore, we continue to investigate strategies for stable transgene expression over multiple generations. Agrobacterium tumefaciens-mediated transient assays work very well in lettuce and are being used to investigate the reactions to pathogen effector proteins. CRISPR Cas-mediated editing also works well and knock-outs are used for gene validation. We are working on allele replacement, sequence addition, and modification of the germline without involving tissue culture.
Another long-term focus of the lab is on downy mildew causing pathogens (DMs). These biotrophic oomycetes cause important diseases in diverse species, but most are under-studied compared to several other pathogens. Our lab uses B. lactucae as a model for understanding the molecular basis for variation in virulence phenotype, fungicide insensitivity, and mating type using genomic sequencing and comparative genomics. These studies have been extended to Peronospora spp., graminicolous DMs that cause major losses in tropical countries, Phytophthora spp., and Pythium uniculatum. A recent finding is that chromosome architecture of diverse species is largely conserved, which enables inferences between species based on synteny.
The latest subject of our crop genomics interests is pistachio. This is an increasingly important crop in California that is also fascinating genetically. It is dioecious, has a small genome, and the predominant rootstocks are F1 seedlings from an interspecific cross between two clonally propagated parents. The millions of rootstocks growing in commercial orchards are possibly the largest set of full sibs on the planet. We have sequenced and assembled the parents and dissected the genetic basis of vigor. We are currently dissecting the genic basis for vigor and sex determination as well as developing methods for transformation and gene editing for pistachio.
All of the above projects have strong bioinformatics components. These involve the acquisition and curation of genetic and sequence data, genome assembly and analysis, gene expression analysis, comparative genomics, and pan genomics, as well as distribution of data to public databases, such as NCBI.
The lab is highly collaborative. We are happy to share our resources and welcome collaborations with both academic and private sector partners, particularly on lettuce, downy mildews, and development of technologies for crop improvement with the goal of increasing global food security.
The lab is also highly diverse. A recent assessment of the lab determined that we could speak 17 different languages and had 12 different passports. We welcome applicants from diverse cultural backgrounds, especially Hispanic communities reflecting the demography of California.
